Probing Gene Expression One Molecule at a Time
Gene expression, a central process to all life, is stochastic because most genes often exist in one or two copies per cell. Although the central dogma of molecular biology has been proven beyond a doubt, due to insufficient sensitivity, stochastic protein production has not been visualized in real-time in an individual cell at the single-molecule level. We report the first direct observation of single protein molecules as they are generated, one at a time in a single live E. coli cell, providing a quantitative description about gene expression.
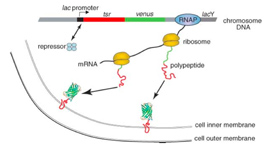
We used a fast maturing fluorescent protein called Venus as a gene expression reporter developed by Mya. We demonstrated a general strategy for measuring live-cell single-molecule fluorophores: detection by localization (Fig. 1) [1]. The key for achieving single molecule detection was to immobilize the fluorescent protein reporter on the cell membrane, by constructing a chimeric fluorescent protein reporter tsr-venus (Fig. 2A), which contains a membrane localization sequence. It is normally difficult to detect single protein molecules inside cytoplasm - their fluorescence is spread by fast diffusion to the entire cell during the image acquisition time, and overwhelmed by the strong cellular autofluorescence. However, molecules on cell membranes diffuse much slower, and therefore can be detected individually with our sensitive microscope. Using this approach, we recorded movies of growing E. coli cells to study the real-time expression from the lac operon in repressed state in real-time (Fig. 2B).
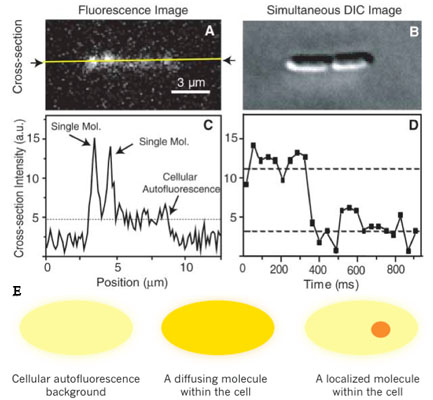
Transcription by RNA polymerase is initiated upon a stochastic dissociation event of the repressor from the operator region of DNA, generating one mRNA molecule. A burst of fusion protein molecules will be synthesized by multiple ribosome molecules bound to the mRNA, yielding fluctuating protein production in time. We observed that under the repressed condition protein molecules are produced in bursts, with each burst originating from a stochastically-transcribed single messenger RNA molecule, and that protein copy numbers in the bursts follow an exponential distribution.These observations were predicted only theoretically previously, accounted for by the competition between mRNA degradation by nuclease and translation by ribosome.
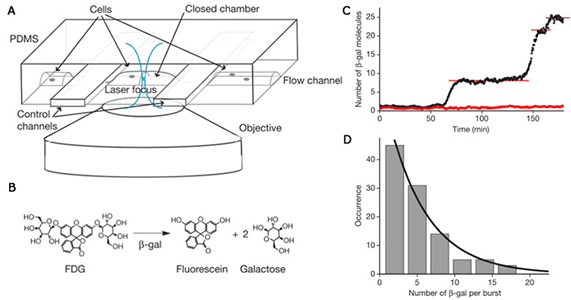
We simultaneously developed a different method using β-glactosidase as a reporter [2] to probe gene expression in living cells with single protein molecule sensitivity: A single copy of β-galactosidase generates many copies of fluorescence molecules when a cell trapped in a microfluidic device was treated with a fluorogenic substrate, resulting in enzymatic amplification in signal and single molecule sensitivity. This technique has been applied to probe gene expression from E. coli as well as individual budding yeast and mouse embryonic stem cells. Again protein production occurs in bursts with exponentially distributed protein copy numbers.
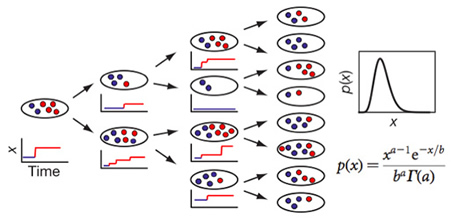
For each gene, the dynamics of the central dogma can be described by two parameters — the burst frequency, a, which is the number of bursts per cell cycle; and the burst size, b, which is the average number of molecules produced per burst. We determined a and b can be from the single-cell time traces, such as in Fig. 3. Under steady-state conditions, temporal fluctuations of gene expression in each cell lineage lead to variation in copy number in an isogenic population of cells. A typical copy-number distribution, which is often asymmetrical, is shown in Fig. 3. A rigorous mathematical relationship between fluctuations in expression and the distribution of protein copy number in a population of cells has been lacking in the literature. A log-normal function has often been used as a convenient phenomenological fit, but it offers no physical insight.
We proved that, under steady-state conditions with uncorrelated and exponentially distributed bursts, the protein copy-number distribution, p(x), can be approximated as a gamma distribution (Fig.4), which has two parameters — a and b,as defined earlier. This allows extraction of intrinsic cellular parameters, a and b from fitting a gamma function to the measured p(x) . At low expression levels, the values for a and b determined in this way are consistent with those derived from the single-cell times traces.
Our single-molecule experiments have provided quantitative descriptions about gene expression in a live cell.
References
[1]Yu, Ji; Xiao, Jie; Ren, Xiaojia; Lao, Kaiqin; Xie, X. Sunney; “Probing gene expression in live cells, one protein molecule at a time,” Science, 311, 1600-1603 (2006).
[2] Cai, Long; Friedman, Nir; Xie, X. Sunney; “Stochastic protein expression in individual cells at the single molecule level,” Nature, 440, 358-362 (2006).
[3] Friedman, Nir; Cai, Long; Xie, X. Sunney “Linking stochastic dynamics to population distribution: An analytical framework of gene expression,“Phys. Rev. Lett.97, 168302 (2006).