Single Molecule Stochastic Event Determines a Cell’s Phenotype
Genetically identical cells in the same environment can exhibit different phenotypes, and a single cell can switch between distinct phenotypes in a stochastic manner. In the classic example of lactose metabolism in E. coli, the lac genes are fully expressed for every cell in a population under high extracellular concentrations of inducers, such as the lactose analog TMG. However, at moderate inducer concentrations, the lac genes are highly expressed in only a fraction of a population, which may confer a fitness advantage for the entire population. Thanks to recent advances on single-molecule imaging, the detailed molecular mechanism that controls the stochastic phenotype switching of a single cell has been revealed.
Lactose metabolism is controlled by the lac operon, which consists of three genes including lactose permease. Expression of the operon is regulated by the lactose repressor, which dissociates from its specific binding sequences of DNA, the lac operators, in the presence of inducers to allow transcription. The production of the permease increases inducer influx, resulting in positive feedback on expression of the lac operon. When the permease is labeled with a yellow fluorescent protein, two different phenotypes are observed in an isogenic population of cells with the fluorescence intensity histogram of the cells exhibiting a bimodal distribution (Fig. 1A & 1B). Interestingly, the low fluorescence cells have a few individual permease molecules, suggesting that one permease molecule is not enough to induce the transition from the low to high fluorescence phenotype. It was determined that 200-300 permease molecules, corresponding to a big burst of gene expression, are needed for this transition.
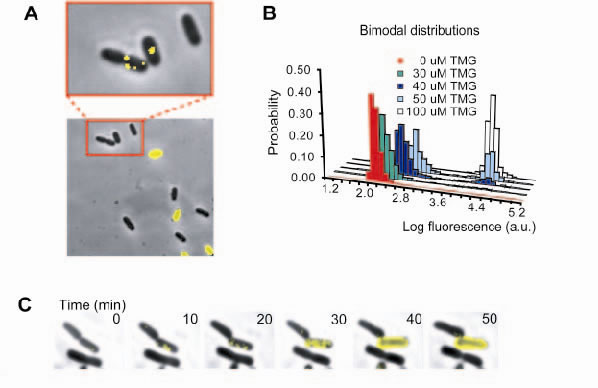
A) In the presence of moderate amounts of inducer (lactose analog TMG), a strain expressing lactose permease fused to yellow fluorescent protein (LacY-YFP) exhibits two phenotypes, all-or-none fluorescence in a fluorescence-phase contrast overlay image. Fluorescence imaging with high sensitivity reveals single molecules of LacY-YFP in the uninduced cells. B) Bimodal fluorescence distributions show that a fraction of the population exists either in an uninduced or induced state, with the relative fractions depending on the inducer concentration. C) A time-lapse sequence captures a phenotype-switching event. One cell switches phenotype to express many LacY-YFP molecules (yellow fluorescence overlay) whereas the other daughter cell does not.

This time-lapse movie shows fluorescence overlay (yellow) on phase contrast images of a cell dividing into two genetically identical daughter cells. One daughter cell changes into a phenotype with very high fluorescence, while the other daughter cell does not. This change in phenotype is the result of the stochastic, full dissociation of the tetrameric repressor from all of its binding sites.
The tetrameric lactose repressor can simultaneously bind to two operators to form a DNA loop. Under low inducer concentrations, the repressor cannot be pulled off the DNA by the inducer; rather, spontaneous, partial dissociations of the repressor result in transcription of one mRNA and a small burst of proteins. However, infrequent events of complete dissociation of the repressor result in large bursts of permease expression (200-300 copies) that trigger induction of the lac operon. This change in phenotype is the result of the stochastic, full dissociation of the tetrameric repressor from all of its binding sites. The time-lapse sequence (Fig.1C) captures such a pheotype–switching event. This illustrates that a stochastic single-molecular event of the complete dissociation of the tetrameric repressor determines a cell’s phenotype, and that a DNA loop is a crucial in maintaining phenotype stability. This is a clear example that a rare event of a single molecule can have significant biological consequences.
Reference
Choi Paul, Cai Long, Frieda Kirsten, Xie Xiaoliang Sunney(2008). A stochastic single-molecule event triggers phenotype switching of a bacterial cell. Science 322:442-446.