Probing Transcription Factor Dynamics
In all kingdoms of life, transcription factors (TFs) regulate gene expression by site specific binding to chromosomal DNA, preventing or promoting the transcription by RNA polymerase. The lac operon of Escherichia coli, a model system for understanding TF-mediated transcriptional control, has been the subject of extensive biochemical, structural, and theoretical studies since the seminal work by Jacob and Monod. However, direct observation of TF mediated gene regulation remains difficult, because TFs exist in small numbers and require single molecule sensitivity. We made the first real-time observation of specific binding of a lac repressor, labeled with a fluorescent protein, to a chromosomal lac operator. We measured the kinetics of binding and dissociation of the repressor in response to metabolic signals [1].
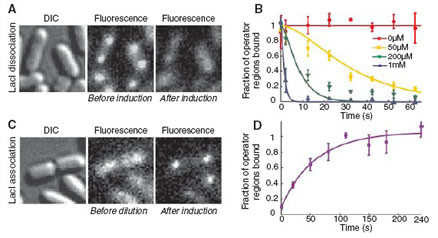
A central question in molecular biology is how a TF molecule or any DNA binding protein searches for specific binding sites on DNA. Target location by TFs is believed to be achieved by facilitated diffusion, in which a TF searches for specific binding sites through a combination of 1D diffusion along a short DNA segment and 3D translocation among DNA segments through cytoplasm [2]. However, direct observation in living cells has not been possible.
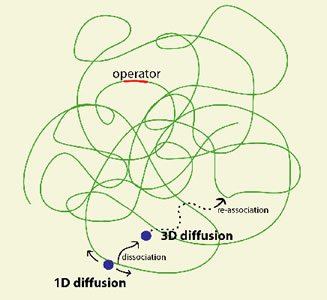
We carried out a live cell study of the search process [1]. Specifically, we characterized the nonspecific binding to DNA, 1D diffusion along DNA segments, and 3D translocation among segments through cytoplasm. We found that in searching for the operator, a lac repressor spends ~90% of time nonspecifically bound to and diffusing along DNA with a residence time of <5 milliseconds. This gives the first quantitative measurement of how individual DNA binding proteins search for their target sequences in a living cell.
References
[1] Elf J, Li GW &Xie XS (2007) Probing transcription factor dynamics at the single-molecule level in a living cell. Science .316 :1191-4 .
[2] Berg, O.G.; Winter, R.B; von Hippel, P.H. “Diffusion-Driven Mechanisms of Protein Translocation on Nucleic Acids. 1. Models and Theory?” Biochemistry 20, 6929 (1981.