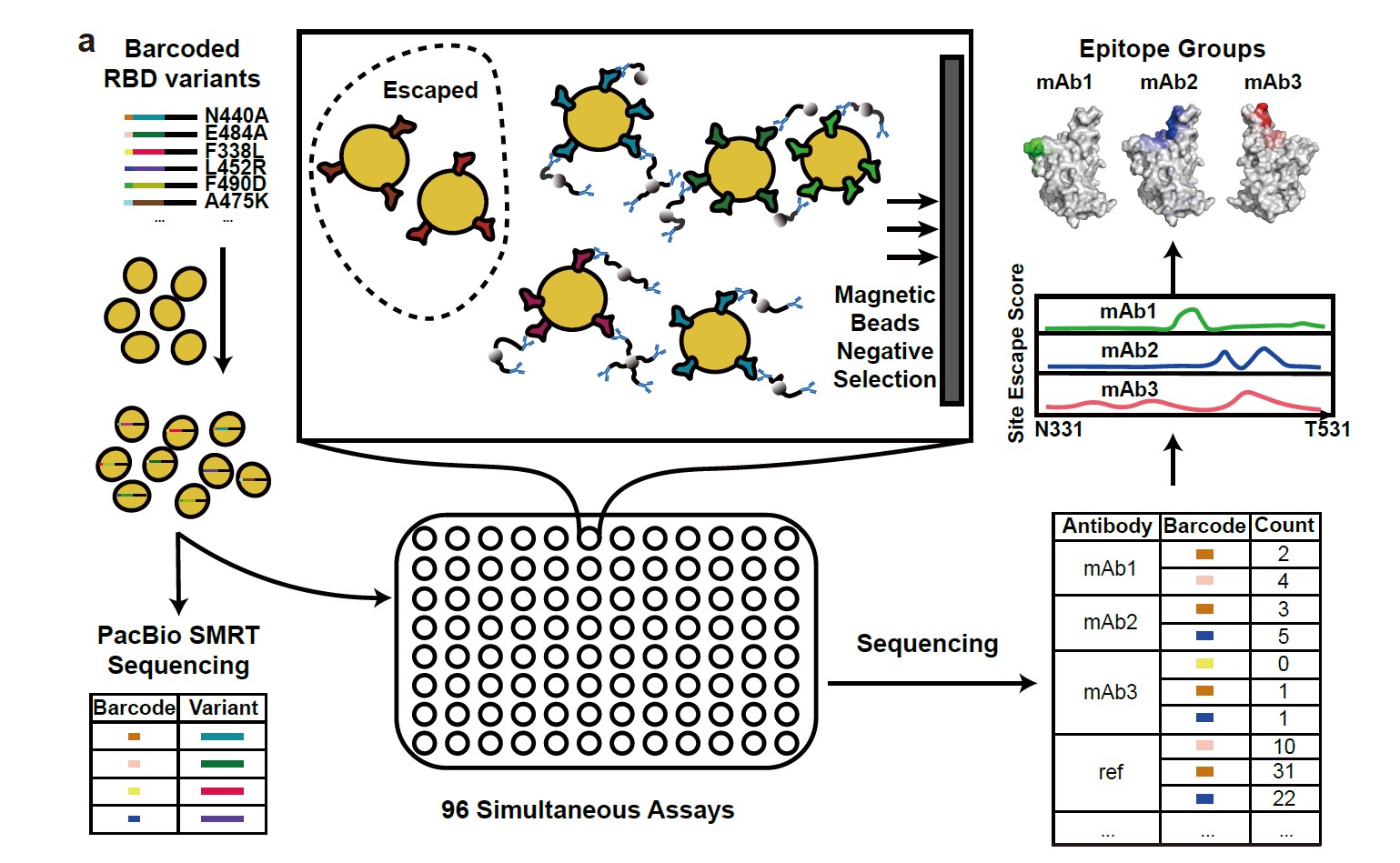
High-throughput neutralizing antibody escape mutation profiling system
The SARS-CoV-2 B.1.1.529 variant (Omicron) contains 15 mutations on the receptor-binding domain (RBD), including G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H. How Omicron would evade RBD neutralizing antibodies (NAbs) requires immediate investigation. A high-throughput yeast display-based antibody escape mutation profiling system was developed and used to determine the RBD escaping mutation profiles for 247 human anti-RBD NAbs and showed that the NAbs could be unsupervised clustered into six epitope groups (Fig.1), which is highly concordant with knowledge-based structural classifications.
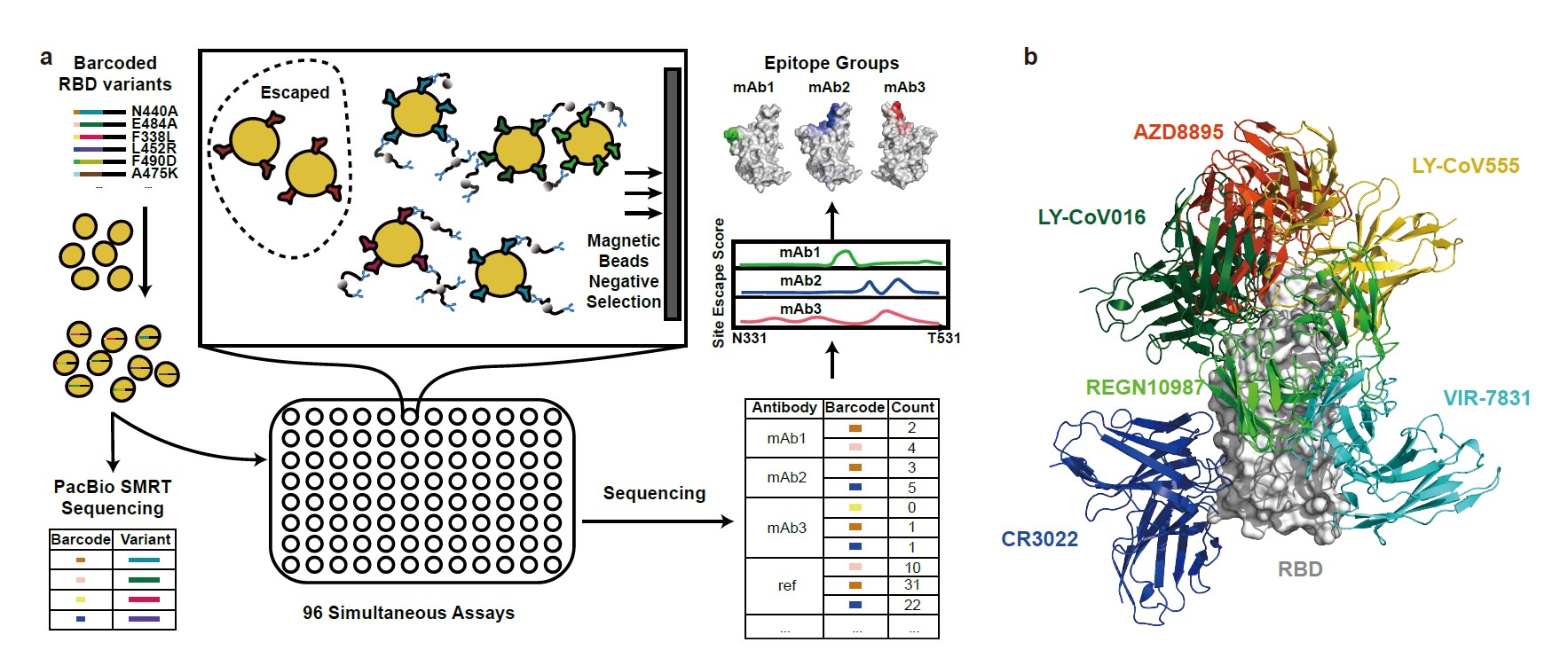
Fig. 1. Omicron greatly reduces the neutralization potency of NAbs of diverse epitopes.
a, Schematic of MACS-based high-throughput yeast display mutation scanning. b, Representative NAb structures of each epitope group. c, t-SNE embedding and unsupervised clustering of SARS-CoV-2 human NAbs based on each antibody escaping mutation profile. A total of 6 epitope groups (Group A-F) could be defined.
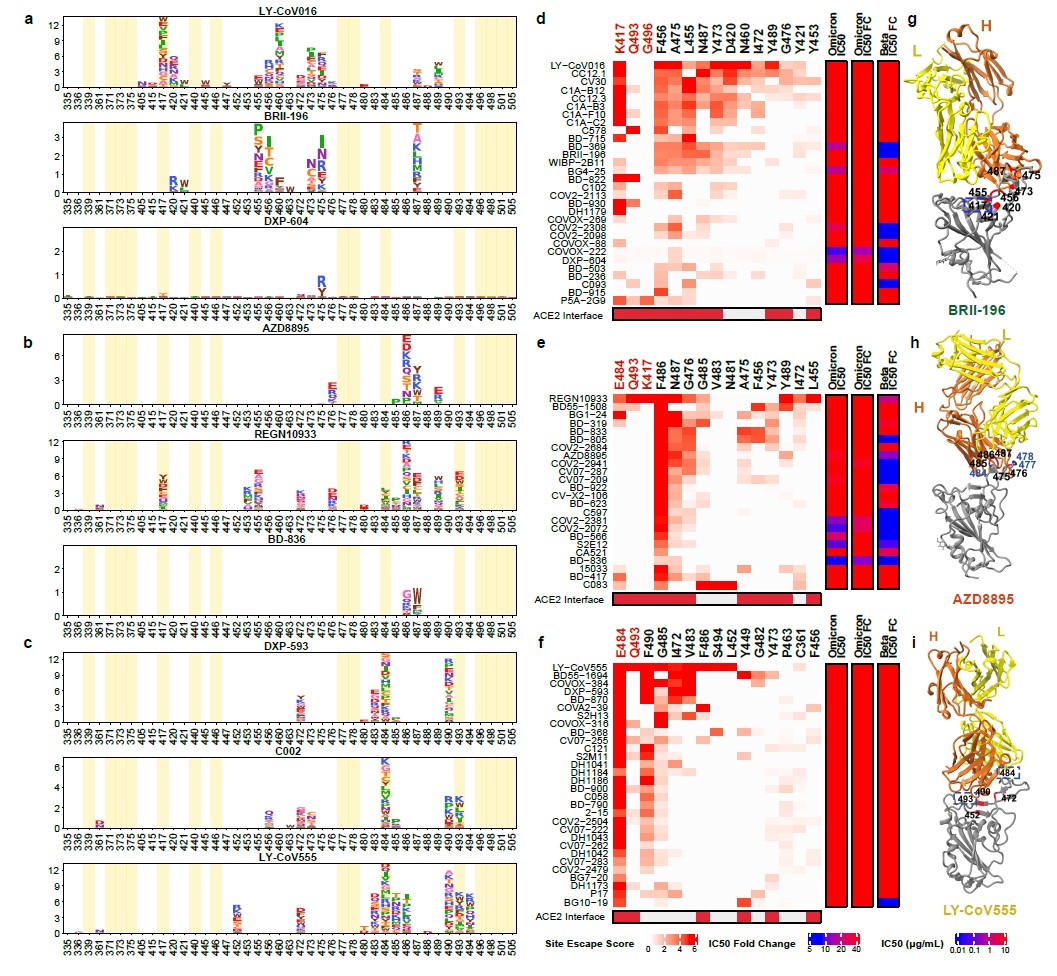
Group A NAbs are often evaded by RBD mutations on K417, D420, F456, A475, L455 sites (Fig. 2d). The NAbs that are insensitive to the K417N single site could also be heavily affected by the combination of K417N and other RBD mutations located on their epitopes of Omicron, causing lost or reduction of neutralization towards Omicron.
Group B NAbs are very sensitive to the change of F486, N487, and G476 (Fig. 2b). Omicron significantly reduced Group B NAbs’ binding affinity to RBD, potentially through S477N/T478K/E484A on their epitope.
Several highly potent antibodies are found in Group C, including BD-368-2/DXP-593, C002, and LY-CoV555. They are mostly prone to the change of E484A mutation in Omicron (Fig. 2f).
Fig. 2. The neutralizing abilities of Group A-C NAbs are mostly abolished by Omicron.
a-c, Escaping mutation profiles of representative NAbs for group A-C, respectively. For each site, the height of a letter indicates the detected mutation escape score of its corresponding residue. Sites mutated in Omicron are highlighted. d-f, Heatmaps of site escape scores for NAbs of epitope group A-C, respectively. ACE2 interface residues are annotated with red blocks, and mutated sites in Omicron are marked red. Annotations on the right side of heatmaps represent pseudovirus neutralizing IC50 fold change (FC) for Omicron and Beta compared to D614G. g-i, Representative structures of group A-C antibodies in complex with RBD. Residues involved in important contacts are labeled. Omicron mutations are marked as blue. NAb escaping mutations (Omicron) inferred from yeast display are labeled with squares.
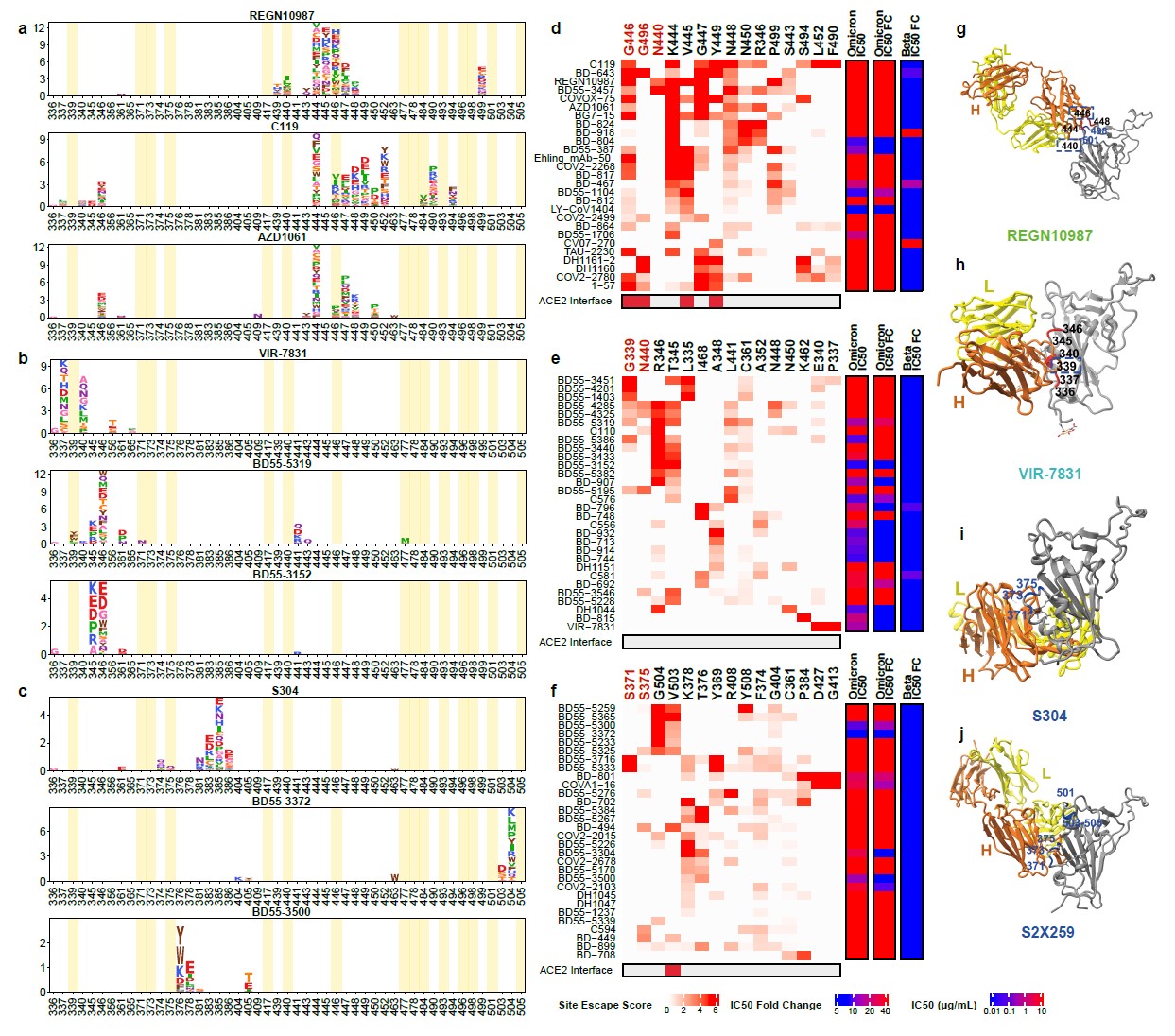
Group D NAbs are sensitive to the changes of N440, K444, G446, and N448(Fig. 3d), resulting in that most Group D NAbs are escaped by Omicron.
Group E NAbs are often sensitive to changes of G339, T345, and R346(Fig. 3b). The G339D mutation would affect a subset of NAbs’ neutralization performance (Fig. 3e). Also, part of Group E NAbs’ epitope would extend to the 440-449 loop, making them sensitive to N440K in Omicron (Fig. 3e).
Group F NAbs are often sensitive to changes of F374, T376, and K378. A part of Group F NAbs is highly sensitive to V503 and G504, similar to the epitopes of S2X259 (Fig. 3f, j), suggesting that they can compete with ACE2. Several NAbs, such as BD55-5300 and BD55-3372, exhibited high neutralization potency compared to other NAbs in Group F (Fig. 3c, 4b). However, their neutralization capability might be undermined by N501Y and Y505H of Omicron (Fig. 3j).
Fig. 3. The majority of Group D-E NAbs are escaped by Omicron.
a-c, Escaping mutation profiles of representative NAbs for group D-E, respectively. For each site, the height of a letter indicates the detected mutation escape score of its corresponding residue. Sites mutated in Omicron are highlighted. d-f, Heatmaps of site escape scores for NAbs of epitope group D-E, respectively. ACE2 interface residues are annotated with red blocks, and mutated sites in Omicron are marked red. Annotations on the right side of heatmaps represent pseudovirus neutralizing IC50 fold change (FC) for Omicron and Beta compared to D614G. g-j, Representative structures of group D-E antibodies in complex with RBD. Residues involved in important contacts are labeled. Omicron mutations are marked as blue. NAb escaping mutations (Omicron) inferred from yeast display are labeled with squares.
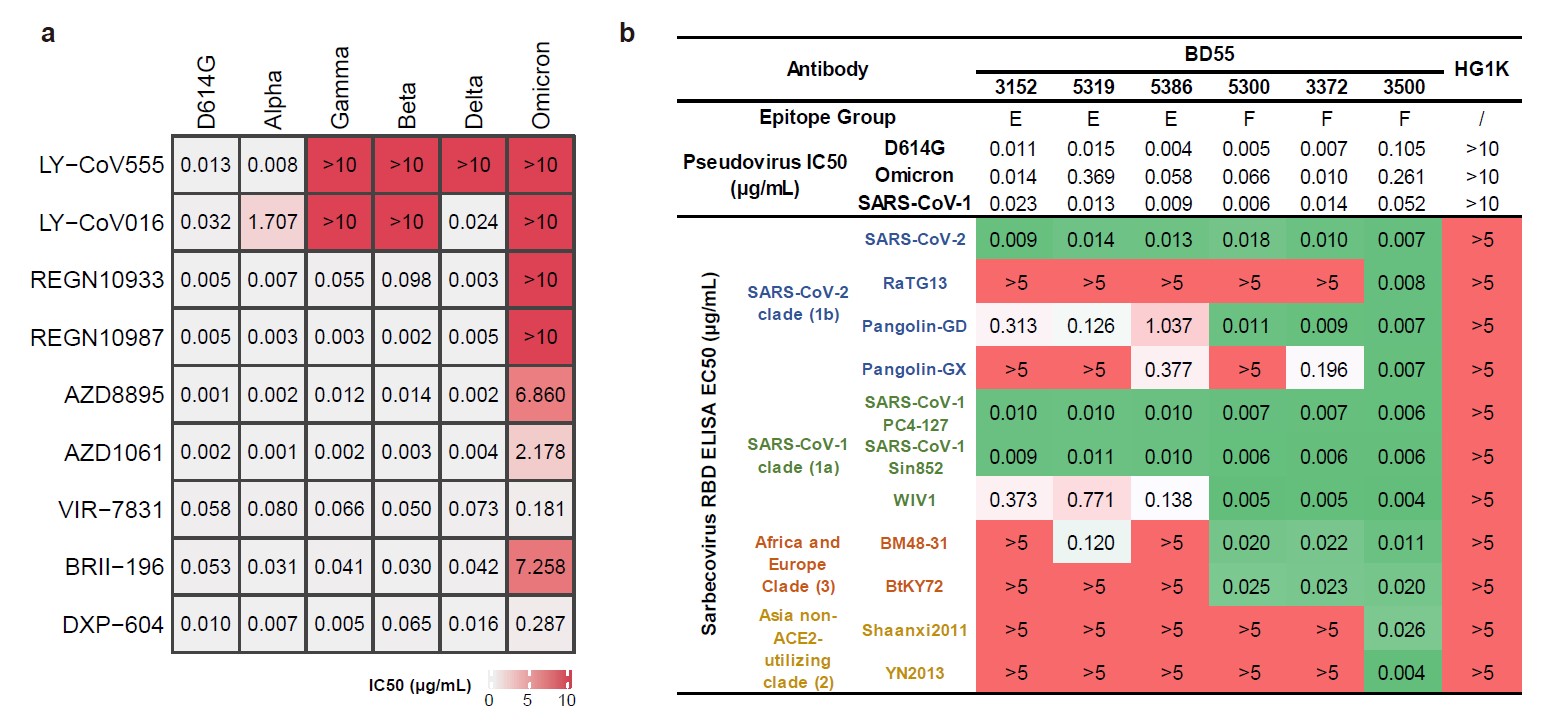
As for NAb drugs, LY-CoV016/LY-CoV555 cocktail, REGEN-10933/REGEN-109876 cocktail, AZD8895, BRII-196, and AZD1061 are escaped by Omicron (Fig. 4a). while VIR-7831 and DXP-604 still function at reduced efficacy. Several NAbs in Group E and F have shown high potency against Omicron and broad pan-sarbecovirus neutralization ability, promising for NAb drug development (Fig. 4b).
Fig. 4. Omicron escapes most NAb drugs.
a, Neutralization of SARS-CoV-2 variants of concern (pseudotyped VSV) by 9 NAb drugs. The pseudovirus neutralization assays for every VOC were performed in biological triplicates. IC50 labeled is the average of three replicates. b, The sarbecovirus neutralization and binding capability of selected potent Omicron-neutralizing antibodies. Monoclonal antibody HG1K (IgG1 antibody against Influenza A virus subtype H7N9) was used as the negative control.
The high-throughput yeast display system developed by us improved the experiment throughput by 1-2 orders of magnitude compared to similar techniques previously reported. In doing so, we could profile the RBD escaping mutation for 247 human anti-RBD NAbs, and what we found offer instructions for developing NAb drugs and vaccines against Omicron and future variants.