SARS-CoV-2 Neutralizing Antibody
The COVID-19 pandemic has spread over the world, and effective therapeutic and prophylactic interventions are urgently needed. Human-sourced monoclonal antibodies generated by convalescent patients’ B cells are promising therapeutic candidates. However, due to VDJ recombination and somatic hypermutation, B cells exhibit diverse B-cell repertoires, and this necessitates the analysis of one B cell at a time to obtain paired immunoglobulin heavy-light chain RNA sequences for monoclonal antibodies production. Using high-throughput single-cell RNA and VDJ sequencing, we rapidly identified multiple SARS-CoV-2 neutralizing antibodies from antigen-binding B cells from convalescent COVID-19 patients (Figure 1).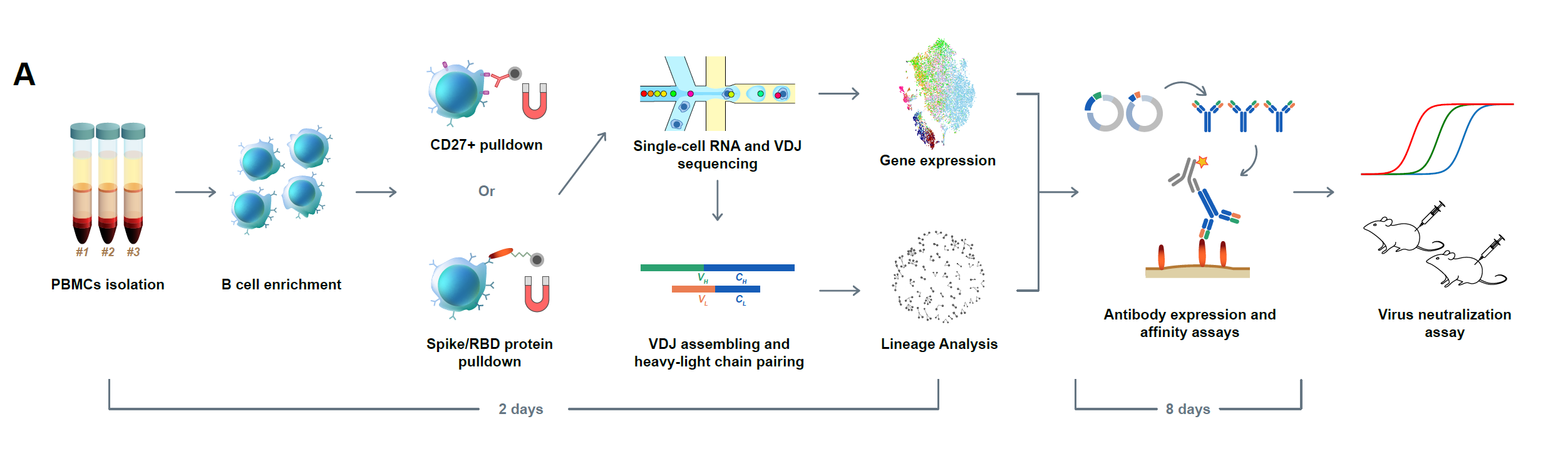
Figure 1. Efficient neutralizing antibody identification through antigen-enriched high-throughput single-cell RNA sequencing. Schematic overview of the neutralizing antibody identification process. The sequence of the mAbs could be obtained within two days using 10X Genomics 5’ VDJ sequencing.
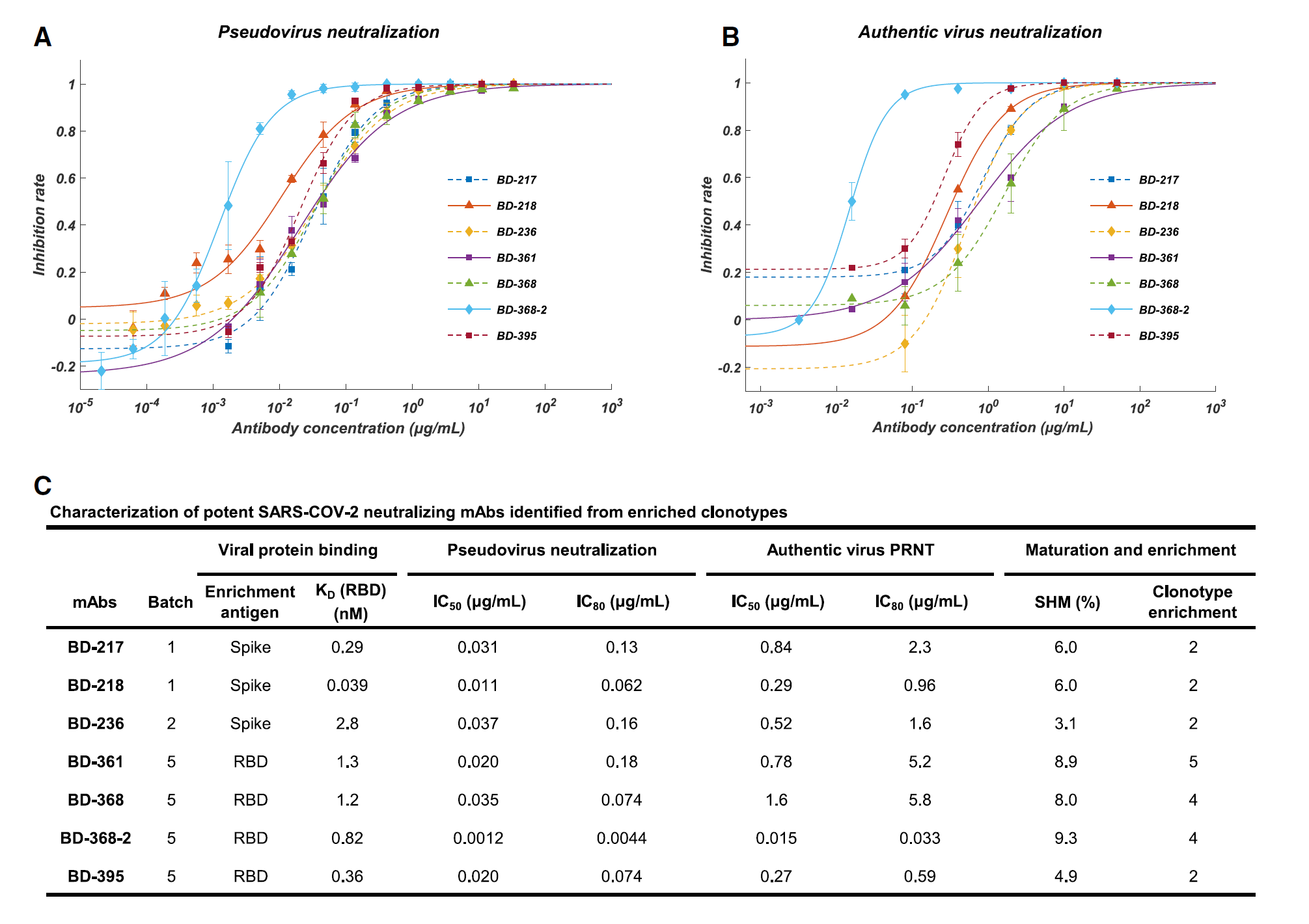
Figure 2. Affinity specificity and neutralizing abilities of the potent neutralizing mAbs. A) Neutralization potency measured by using a pseudotyped virus neutralization assay. B) Neutralization potency measured by an authentic SARS-CoV-2 plaque reduction neutralizing test (PRNT) assay. C) Characteristics of the neutralizing mAbs. IC50 and IC80 were calculated by using a four-parameter logistic curve-fitting. Kd targeting RBD was measured by using SPR with a 1:1 binding model.
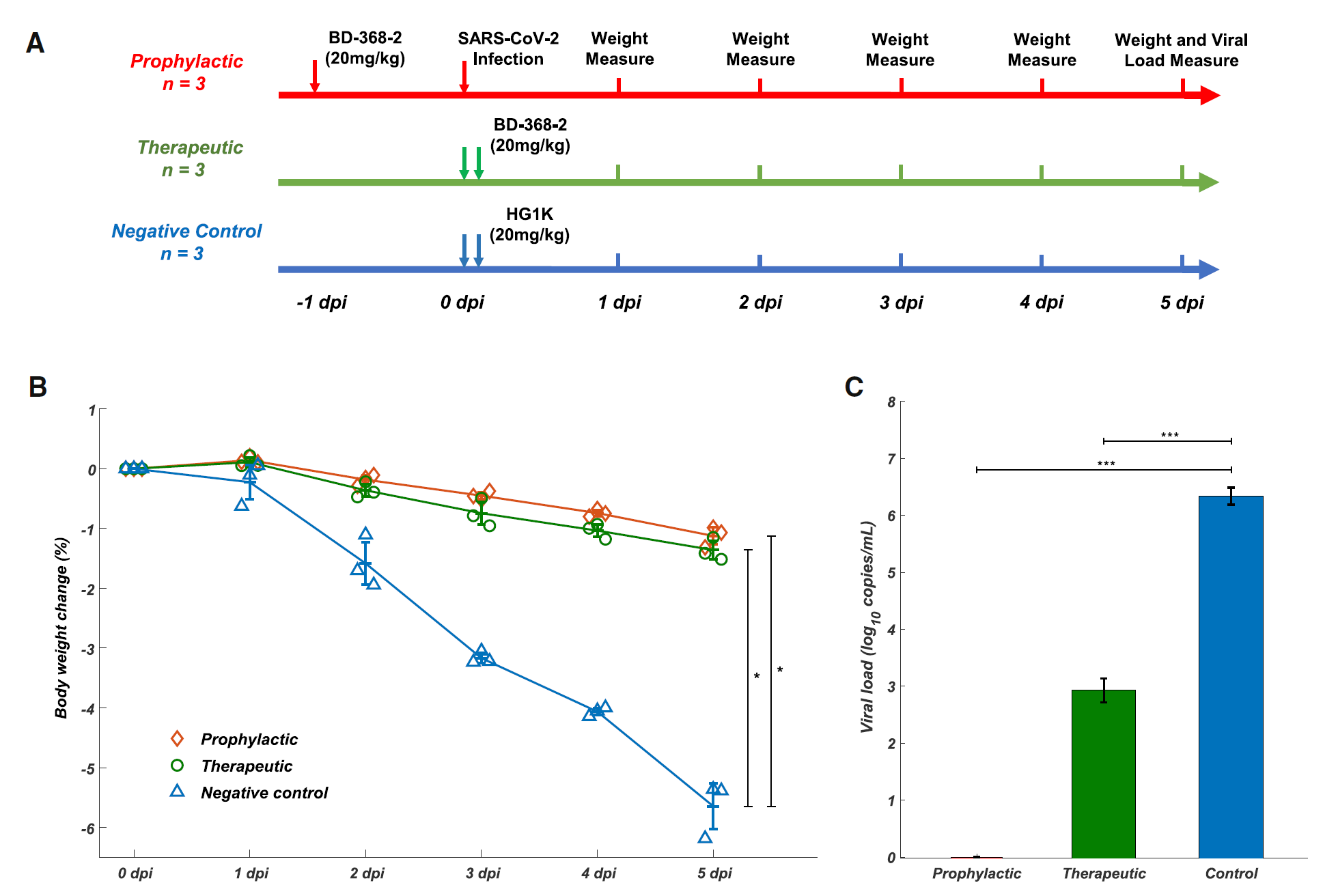
Figure 3. BD-368-2 showed high therapeutic and prophylactic efficacy in SARS-CoV-2-infected hACE2 transgenic mice. A) Experimental design for therapeutic and prophylactic testing of BD-368-2 in hACE2 transgenic mice. BD-368-2 or unrelated antibody HG1K (20 mg/kg of body weight) was intraperitoneally injected into the transgenic mice before or after SAR-CoV-2 infection. B) Body weight change (%) of the hACE2 transgenic mice recorded over 5 days (one-sided permutation test, *p<0.05). Each group contains 3 mice. C) Virus titers of lung tissue at 5 dpi. The viral loads of the lung were determined by qRT-PCR (one-tailed t-test, ***p<0.001).
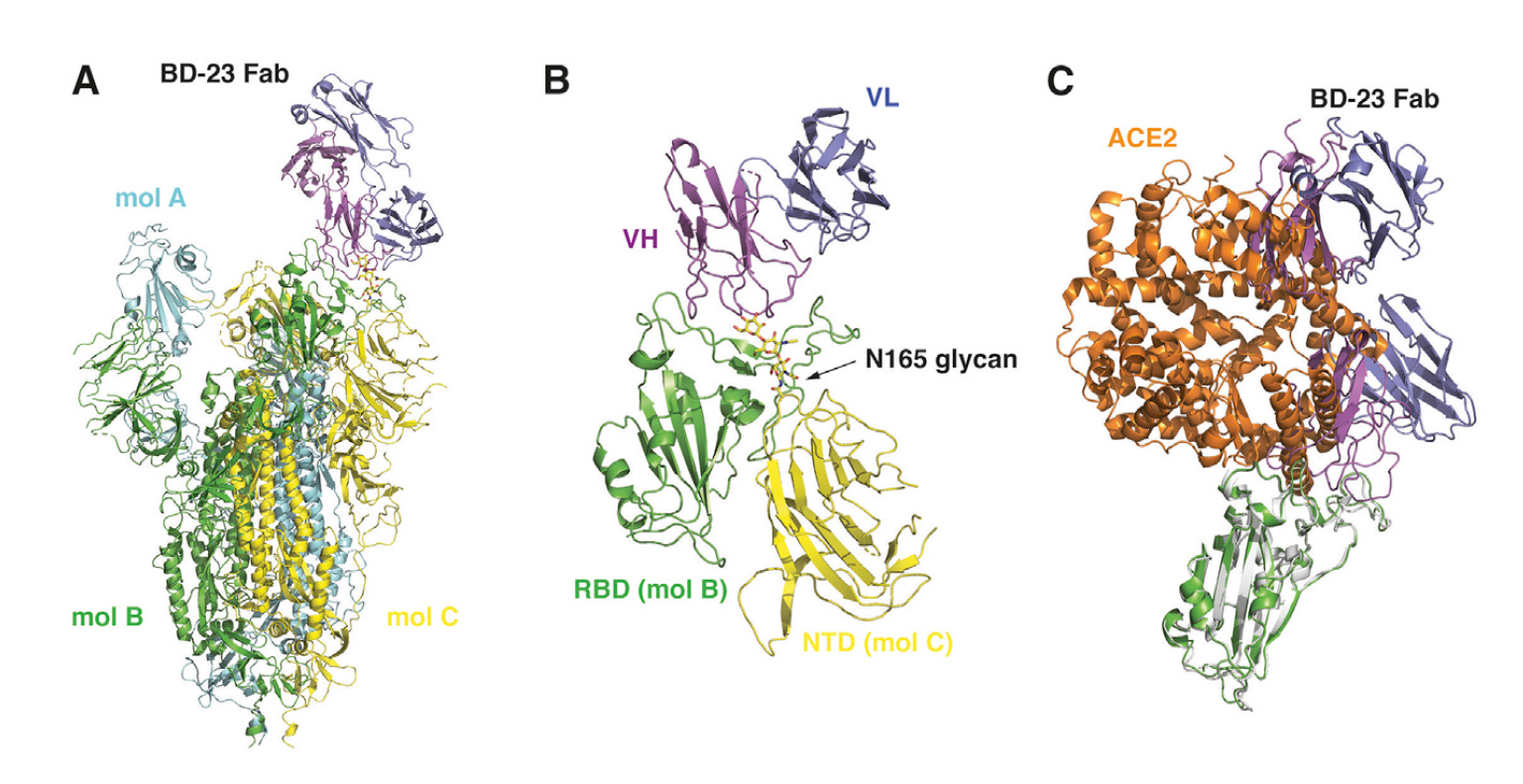
Figure 4. Cryo-EM structure of BD23-Fab in complex with the Spike trimer. A) Cryo-EM structure of the S trimer in complex with BD23-Fab reconstructed at 3.8 Å resolution. The three protomers in the S trimer are depicted in cyan, green, and yellow, respectively. BD23-Fab is depicted in magenta (heavy chain) and blue (light chain). B) N165 glycan in the NTD of protomer C facilitates the interaction between BD23-Fab and the RBD of protomer B. C) The crystal structure of the RBD/ACE2 complex is overlaid onto the RBD/BD23-Fab structure. BD23-Fab would collide with ACE2 and therefore block the interaction between RBD and ACE2. RBD is shown in green and white, whereas ACE2 in orange.
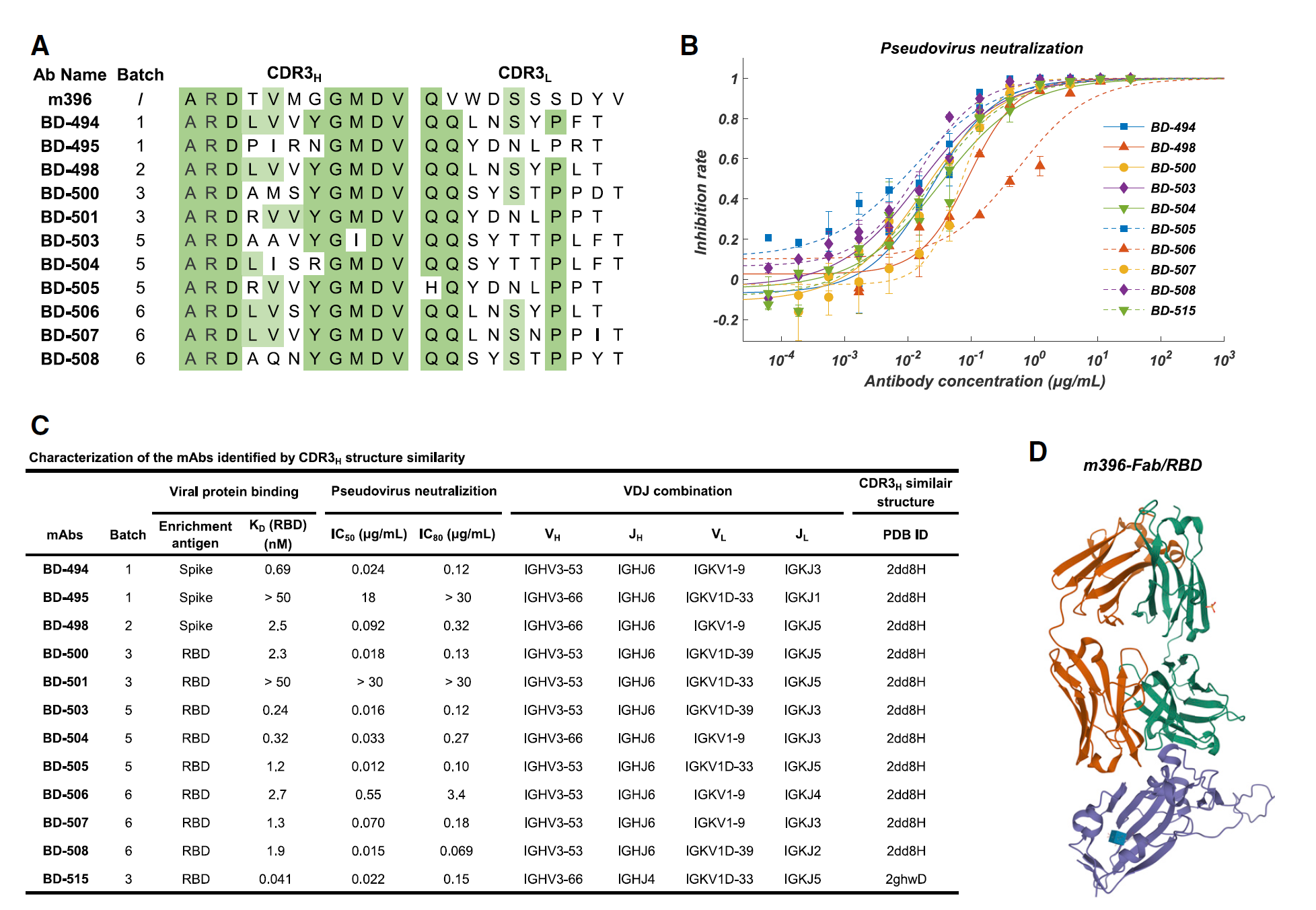
Figure 5. Characteristics of the neutralizing mAbs identified based on CDR3H structural similarity to SARS-CoV neutralizing mAbs. A) The CDR3 sequence comparison between SARS-CoV neutralizing mAb m396 and the SARS-CoV-2 neutralizing mAbs identified based on CDR3H structure similarity. B) Neutralization potency measured by using a pseudotyped virus neutralization assay. C) Characteristics of the neutralizing mAbs identified based on structure similarity. The CDR3H structure prediction was performed using FREAD. D) The crystal structure of m396-Fab/SARS-CoV-RBD.