Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants

On August 25, 2021, the research group of Xie Xiaoliang and Xiao Junyu jointly published a paper entitled “Structures of SARS-CoV-2 B.1.351 neutralizing antibodies provide insights into cocktail design against concerning variants” on Cell Research.
In the previous study, our team discovered a set of monoclonal antibodies that can effectively neutralize the B.1.351 mutant by analyzing the immune responses of long-term convalescents and vaccine recipients to the B.1.351 mutant (Cao et al. , Cell Research 31:732-741, 2021). This work further analyzes the molecular mechanism and pairing of these neutralizing antibodies.
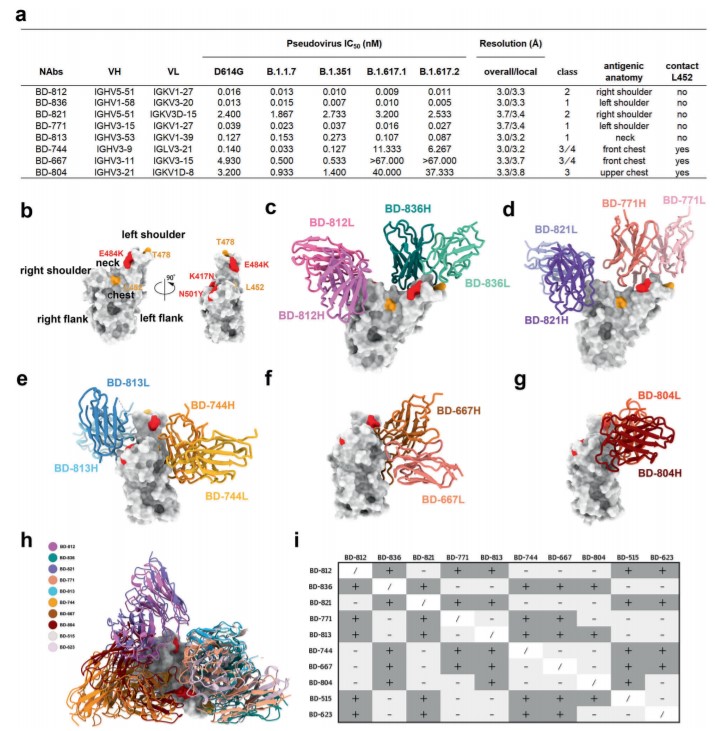
First, through the pseudovirus neutralization experiment, it was found that the two antibodies of BD-812 and BD-836 had very high activity, and the neutralization of the B.1.351 pseudovirus was close to the pM level. High-resolution single-particle cryo-electron microscopy (cryo-EM) studies have shown that BD-812 and BD-836 bind to the left and right shoulders of the receptor binding domain (RBD) of the spike protein of the B.1.351 mutant strain, respectively. BD-812 and BD-836 have binding epitopes that do not interfere with each other, and can directly hinder the binding of RBD to ACE2 receptor. It is particularly important that the binding of BD-812 and BD-836 to RBD is not affected by the two mutation sites of L452R and T478K in the Delta mutant, and the pseudovirus experiments also show that they can effectively neutralized the B.1.617.2 variant.
The work also analyzes the mechanism by which other antibodies target the B.1.351 spike protein. The combination of BD-821 and BD-771 is very similar to the above-mentioned BD-812/BD-836 binding mode, and both also have potent neutralizing activity against the Delta mutant. But BD-821 covered a smaller interface on the RBD compared to BD-812, explaining its weaker neutralizing activity. BD-813 binds to the back of the RBD and can directly block the binding to ACE2, and also maintains activity against the Delta mutant. BD-744, BD-667 and BD-804 bind to the “chest”of RBD and do not overlap the binding epitope of ACE2, but the epitopes of these antibodies all involve the Leu452 site,and the Delta mutant had a severe escape against them. Clearly, the L452R mutation in the Delta mutant would render many neutralizing antibodies targeting this region ineffective.
In conclusion, this study systematically analyzed a series of fully humanized neutralizing antibodies targeting the B.1.351 spike protein, and analyzed their combination strategies and responses to the Delta mutant based on structural information. In particular, the BD-812/BD-836 antibody combination from long-term convalescents performed very well, showing pM-level activity against the B.1.617.1 and B.1.617.2 variants,,and is expected to become a candidate drug for SARS-CoV-2 VOCs.